A First Attempt at Executable Packaging with pex
[ ]For an ongoing project here at work, I’ve written some Python tooling for automatically importing some chromatograms generated by the instrument software and extracting peak data from them. This workflow is necessary because our instrument is old, and the software that drives it was never actually intended to generate chromatograms: the only way to get the time scan data out is to … print to PDF.
Yes, really.
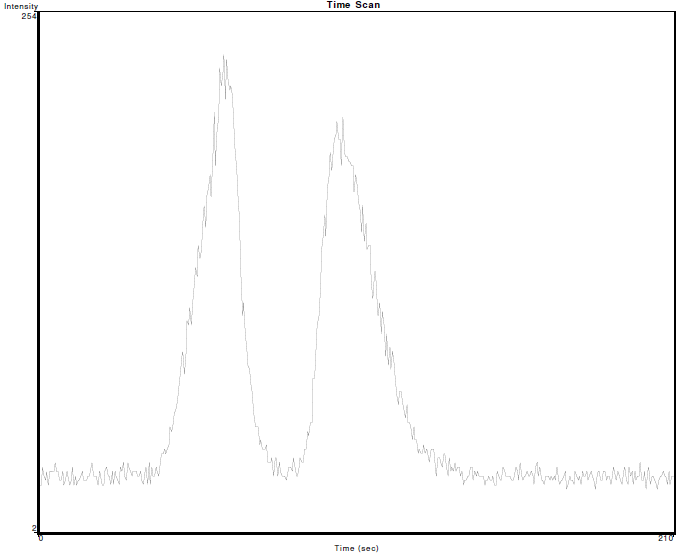
So, the data (which is the intensity of the emission signal for chromium over time measured from an ICP-OES) looks like this:
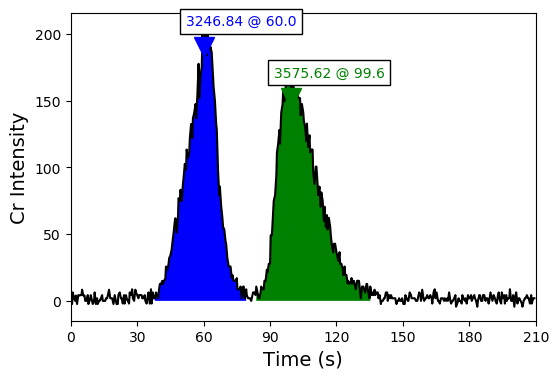
And what we want, ultimately, is for it to look like this:
Also, we want the peak and trace information exported to Excel-readable formats in various ways, for downstream analysis.
Skipping directly to the end of the development story (the full telling of which must wait for another post):
it works! Through a combination of
numpy ![]() ,
,
scipy ![]() ,
,
peakutils ![]() ,
and a nifty PDF-inspecting library called
,
and a nifty PDF-inspecting library called minecart ![]() ,
I can pull what I need out of the PDFs, detect the peaks in the data trace, and calculate
(what are hopefully) all of the properties of the peaks that we’ll need.
Then, the plotting niftiness of
,
I can pull what I need out of the PDFs, detect the peaks in the data trace, and calculate
(what are hopefully) all of the properties of the peaks that we’ll need.
Then, the plotting niftiness of
matplotlib ![]() and the XLSX export capabilities of
and the XLSX export capabilities of openpyxl ![]() work nicely to generate the outputs we need.
work nicely to generate the outputs we need.
With the working code in hand, though, the question presents itself: How to enable other people,
the ones who will actually be collecting the data, to use this tool? Well, happily,
the fine folks over at Talk Python ![]() and Python Bytes
and Python Bytes ![]() had
made mention
a few times
of the tool
had
made mention
a few times
of the tool pex ![]() (see also here
and here),
which can create (almost) single-object Python executables.
Sounded like a great first thing to try.
(see also here
and here),
which can create (almost) single-object Python executables.
Sounded like a great first thing to try.
pex has a lot of options, and a lot of different ways you can go about hooking into the
code you compile into the .pex file. What I’m describing here is
what worked for me on this first attempt to use it.
The first step was to pull the code out of Jupyter and set it up as a proper Python package,
which I called icicp. (The data being analyzed is from an
ICP-OES method
with an ion chromatography
pre-separation; thus, IC-ICP-OES, and thus icicp.)
Having built packages a few times before, it was a pretty straightforward process.
pex allows you to link to a default entry point when you build the icicp.pex, so that
it can just be invoked as python icicp.pex to automatically execute the desired chain
of code. (Note that you DON’T have to specify this entry point in your setup.py
for pex to be able to use it.)
I’d converted the Jupyter notebook cells responsible for actually driving the
workhorse code into a runner.py helper module; a main() entry point function in that
module worked nicely as the target for this default .pex execution.
So, at the point of being ready to start configuring pex, my package tree looked like this:
icicp/
|-- icicp/
| |-- __init__.py
| | : __version__
| |-- chroma.py
| |-- core.py
| |-- peak.py
| |-- runner.py
| : main()
|-- setup.py
|-- requirements-pex.txt
Since this code isn’t currently meant for public distribution
(too inflexible and application-specific),
a minimal setup.py sufficed:
from setuptools import setup
from icicp import __version__
setup(
name="icicp",
version=__version__,
description="IC-ICP-OES Chromatogram Analyzer",
packages=["icicp"],
python_requires=">=3.6",
)
AFAICT, pex doesn’t care if you’re running it inside a virtual environment, or within
the root directory of the source tree of a package on disk, or wherever—no matter what, it doesn’t
inspect its environment to see if there might be code you want it to include. It stuffs
exactly the packages you tell it to into the built .pex file, and ONLY those packages
(plus any dependencies).
I haven’t yet figured out how to tell pex to include an on-disk source tree, so
I had to make a wheel of the icicp package (python setup.py bdist_wheel).
For the other requirements, I specified pinned versions for my top-level dependencies
in requirements-pex.txt:
attrs==18.1
numpy==1.15
matplotlib==2.1.2
minecart==0.3.0
openpyxl==2.5.9
peakutils==1.3.0
scipy==1.1
tqdm==4.28.1
After a pip install pex in my working virtual environment, the following command
(Windows 10 environment) successfully built a working icicp.pex:
C:\...\icicp> pex -v -r requirements-pex.txt dist\icicp-0.1.3-py3-none-any.whl -e icicp.runner:main -o icicp.pex
icicp==0.1.3 -> icicp 0.1.3
attrs==18.1 -> attrs 18.1.0
numpy==1.15 -> numpy 1.15.0
matplotlib==2.1.2 -> matplotlib 2.1.2
minecart==0.3.0 -> minecart 0.3.0
openpyxl==2.5.9 -> openpyxl 2.5.9
peakutils==1.3.0 -> PeakUtils 1.3.0
scipy==1.1 -> scipy 1.1.0
tqdm==4.28.1 -> tqdm 4.28.1
cycler>=0.10 -> cycler 0.10.0
pytz -> pytz 2018.7
python-dateutil>=2.1 -> python-dateutil 2.7.5
six>=1.10 -> six 1.11.0
pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 -> pyparsing 2.3.0
pdfminer3k -> pdfminer3k 1.3.1
et-xmlfile -> et-xmlfile 1.0.1
jdcal -> jdcal 1.4
ply>=3.4 -> ply 3.11
pytest>=2.0 -> pytest 4.0.1
atomicwrites>=1.0 -> atomicwrites 1.2.1
py>=1.5.0 -> py 1.7.0
pluggy>=0.7 -> pluggy 0.8.0
more-itertools>=4.0.0 -> more-itertools 4.3.0
colorama; sys_platform == "win32" -> colorama 0.4.1
setuptools -> setuptools 40.6.2
pex: Building pex: 79813.3ms
pex: Resolving distributions: 79804.3ms
Saving PEX file to icicp.pex
BEWARE: pex caches packages, apparently including local wheels. Thus, using the
approach of building icicp into a wheel, I had to bump the version number inside
__init__.py any time I made changes to the icicp code, or else pex would use the
cached version and my changes wouldn’t get incorporated into icicp.pex.
The pex cache is stored by default in ~\.pex\; presumably deleting the relevant
wheels here would have worked to pull in the revised code.
Alternatively, pex does expose a --disable-cache option, but this might result in
all dependencies being re-downloaded unnecessarily, since I don’t know whether
pex checks pip’s cache before reaching out to PyPI.
While .pex files can be made executable
fairly straightforwardly on Linux,
it’s not so simply done on Windows. Rather than trying to associate the .pex extension with
Python, I went the route of creating a simple launch script, icicp.bat:
@echo off
echo Unpacking and running IC-ICP data workup tool...
call python icicp.pex
pause
As long as a suitable system Python version is available, when placed
in the same directory as icicp.pex, double-clicking this script kicks off the
internal code using icicp.runner:main() as the point of entry.
The way the data analysis code is set up, all the user needs to do is drop
the PDFs they want analyzed into the same folder and run the script:
Unpacking and running IC-ICP data workup tool...
Collecting list of PDF files...
100%|############| 23/23 [00:00<?, ?it/s]
Importing chromatogram data from PDFs...
100%|############| 16/16 [00:00<00:00, 32.25it/s]
Generating image and Excel outputs for each PDF...
100%|############| 16/16 [00:04<00:00, 3.36it/s]
Generating summary Excel file for all PDFs...
...Done!
Output stored in 'output_20181204_131957'
Press any key to continue . . .
The progress bars in the console output were generated by the
always-handy tqdm ![]() .
.